|
Available
Oxford Centre for Soft and Biological Matter
|
DNA self-assembly
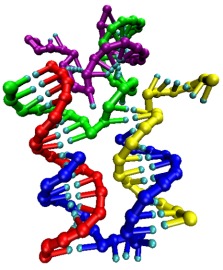 DNA is an ideal material for nanoengineering, because the specificity of the
base pairing allows target structures to be encoded in the base sequences of
component oligonucleotides.
For example, the group of Professor Andrew Turberfield in the Department of Physics in Oxford are able
to self assemble nanostructures, such as a DNA tetrahedron, simply by cooling
down mixtures of appropriately designed oligonucleotides. They have also
developed DNA nanodevices, such as DNA "nanotweezers" (illustrated right, in the
process of opening) and
a DNA walker.
DNA is an ideal material for nanoengineering, because the specificity of the
base pairing allows target structures to be encoded in the base sequences of
component oligonucleotides.
For example, the group of Professor Andrew Turberfield in the Department of Physics in Oxford are able
to self assemble nanostructures, such as a DNA tetrahedron, simply by cooling
down mixtures of appropriately designed oligonucleotides. They have also
developed DNA nanodevices, such as DNA "nanotweezers" (illustrated right, in the
process of opening) and
a DNA walker.
DNA is able to exist in a number of forms, as well as the standard B-form; for example, Z-DNA that has a left-handed helix, and the four-stranded G-quadruplexes. These forms are biologically relevant, since certain sequences in the genome are known to have a propensity to form these structures, and transitions between the different forms may play a role in gene regulation.
We have now developed a coarse-grained models of DNA that allows us to simulate DNA on the time scales associated with self-assembly and structural transitions. As well as applying it to visualize the mechanisms of self-assembly and action of nanodevices, we also wish to extend the model to apply it in a biological context.
This project is in collaboration with the group of Dr Ard Louis in The Rudolf Peierls Centre for Theoretical Physics.
Relevant Publications-
T.E. Ouldridge, A.A. Louis and J.P.K. Doye, J. Chem. Phys, 134, 085101 (2011)
Structural, mechanical and thermodynamic properties of a coarse-grained DNA model -
T.E. Ouldridge, A.A. Louis and J.P.K. Doye, Phys. Rev. Lett. 104, 178101
(2010)
DNA nanotweezers studied with a coarse-grained model of DNA -
T.E. Ouldridge, I.G. Johnston, A.A. Louis and J.P.K. Doye, J. Chem. Phys.
130, 065101 (2009)
The self-assembly of DNA Holliday junctions studied with a minimal model