|
|
Characterizing the free-energy landscapes of DNA origamis
Chak Kui Wong, Chuyan Tang, John S. Schreck, Jonathan P.K. Doye
Nanoscale 14, 2638-2648 (2022)
Abstract
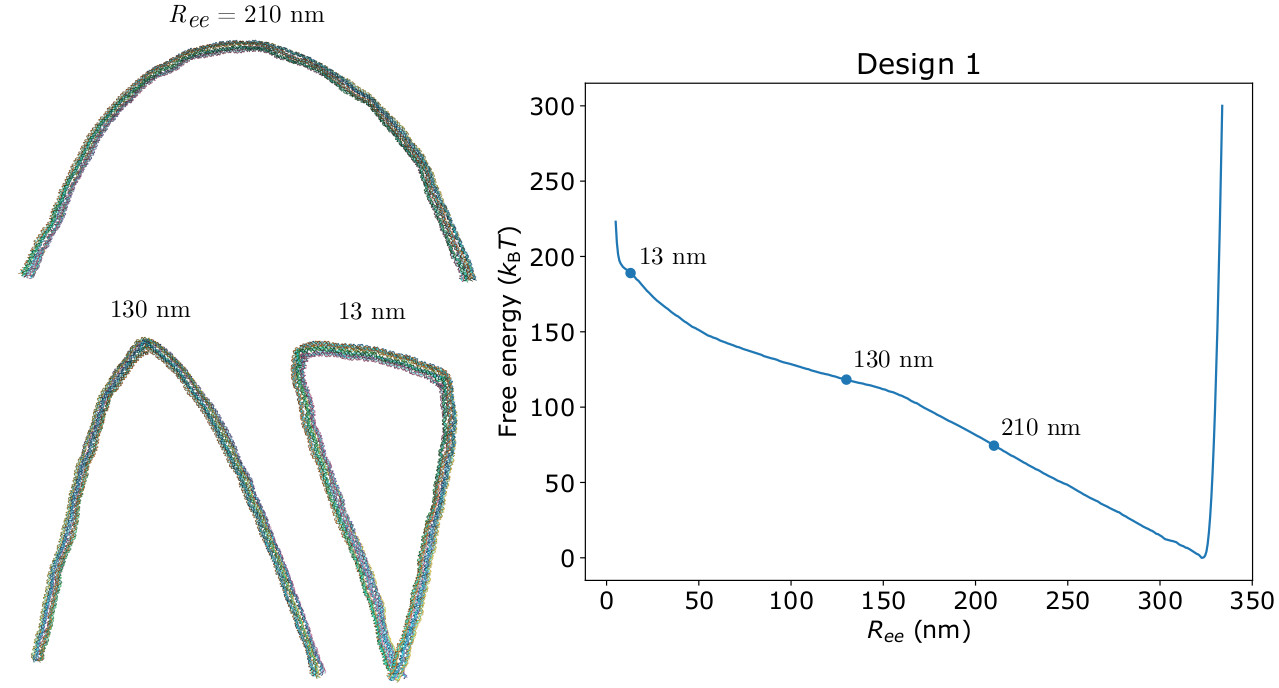
We show how coarse-grained modelling combined with umbrella sampling using distance-based order parameters can be applied to compute the free-energy landscapes associated with mechanical deformations of large DNA nanostructures. We illustrate this approach for the strong bending of DNA nanotubes and the potentially bistable landscape of twisted DNA origami sheets. The homogeneous bending of the DNA nanotubes is well described by the worm-like chain model; for more extreme bending the nanotubes reversibly buckle with the bending deformations localized at one or two ``kinks''. For a twisted one-layer DNA origami, the twist is coupled to the bending of the sheet giving rise to a free-energy landscape that has two nearly-degenerate minima that have opposite curvatures. By contrast, for a two-layer origami, the increased stiffness with respect to bending leads to a landscape with a single free-energy minimum that has a saddle-like geometry. The ability to compute such landscapes is likely to be particularly useful for DNA mechanotechnology and for understanding stress accumulation during the self-assembly of origamis into higher-order structures.The full paper is available from Nanoscale and arXiv.
The paper was selected for the 2022 Nanoscale HOT Article Collection