|
|
Designer protein assemblies with tunable phase diagrams in living cells
Meta Heidenreich, Joseph M. Georgeson, Emanuele Locatelli, Lorenzo Rovigatti, Saroj Kumar Nandi, Avital Steinberg, Yotam Nadav, Eyal Shimoni, Samuel A. Safran, Jonathan P. K. Doye, Emmanuel D. Levy
Nat. Chem. Biol. 16, 939-945 (2020)
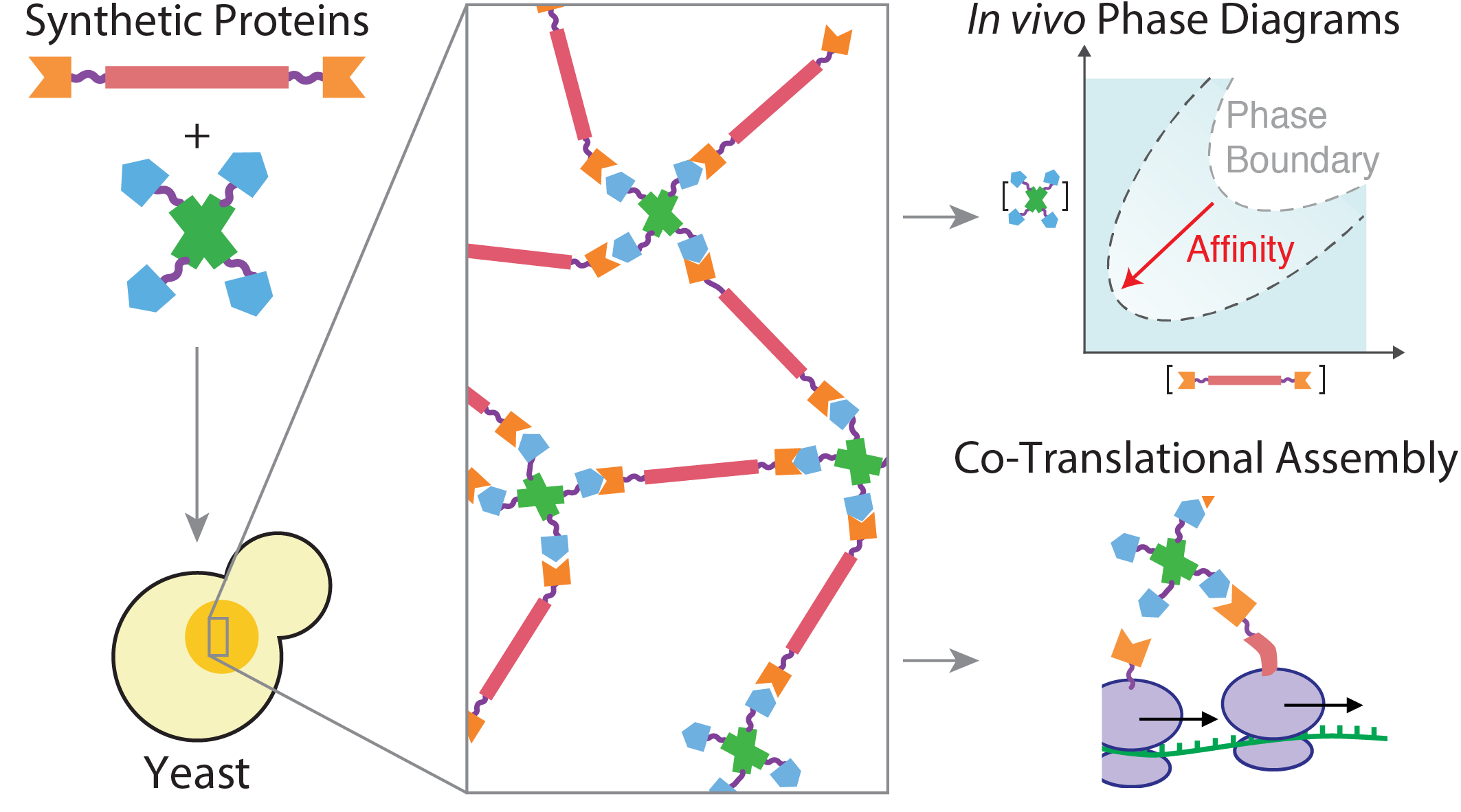
Abstract
The self-organization of proteins into specific assemblies is a hallmark of biological systems. Principles governing protein-protein interactions have long been known. However, principles by which such nanoscale interactions generate diverse phenotypes of mesoscale assemblies, including phase-separated compartments, remains challenging to characterize and understand. To illuminate such principles, we create a system of two proteins designed to interact and form mesh-like assemblies in living cells. We devise a strategy to map high-resolution phase diagrams of the system in vivo, thus revealing its mesoscale self-assembly signatures. The structural modularity of the two protein components allows straightforward modification of their molecular properties, enabling us to characterize how point mutations that change their interaction affinity impact the phase diagram and material state of the assemblies in vivo. Both, the phase diagrams and their dependence on interaction affinity were captured by theory and simulations, including out-of-equilibrium effects seen in growing cells. Applying our system to interrogate biological mechanisms of self-assembly, we find that co-translational protein binding suffices to recruit an mRNA to the designed micron-scale structures.The full paper is available from Nature Chemical Biology and bioRxiv.
The paper was highlighted in a Nature Chemical Biology News & Views feature by Roshan Regy and Jeetain Mittal.
It was also recommended by Shoshana Wodak in Faculty Opinions.