|
|
Coarse-grained modelling of the structural properties of DNA Origami
Benedict K. Snodin, John S. Schreck, Flavio Romano, Ard A. Louis and Jonathan P.K. Doye
Nucleic Acids Res. 47, 1585-1597 (2019)
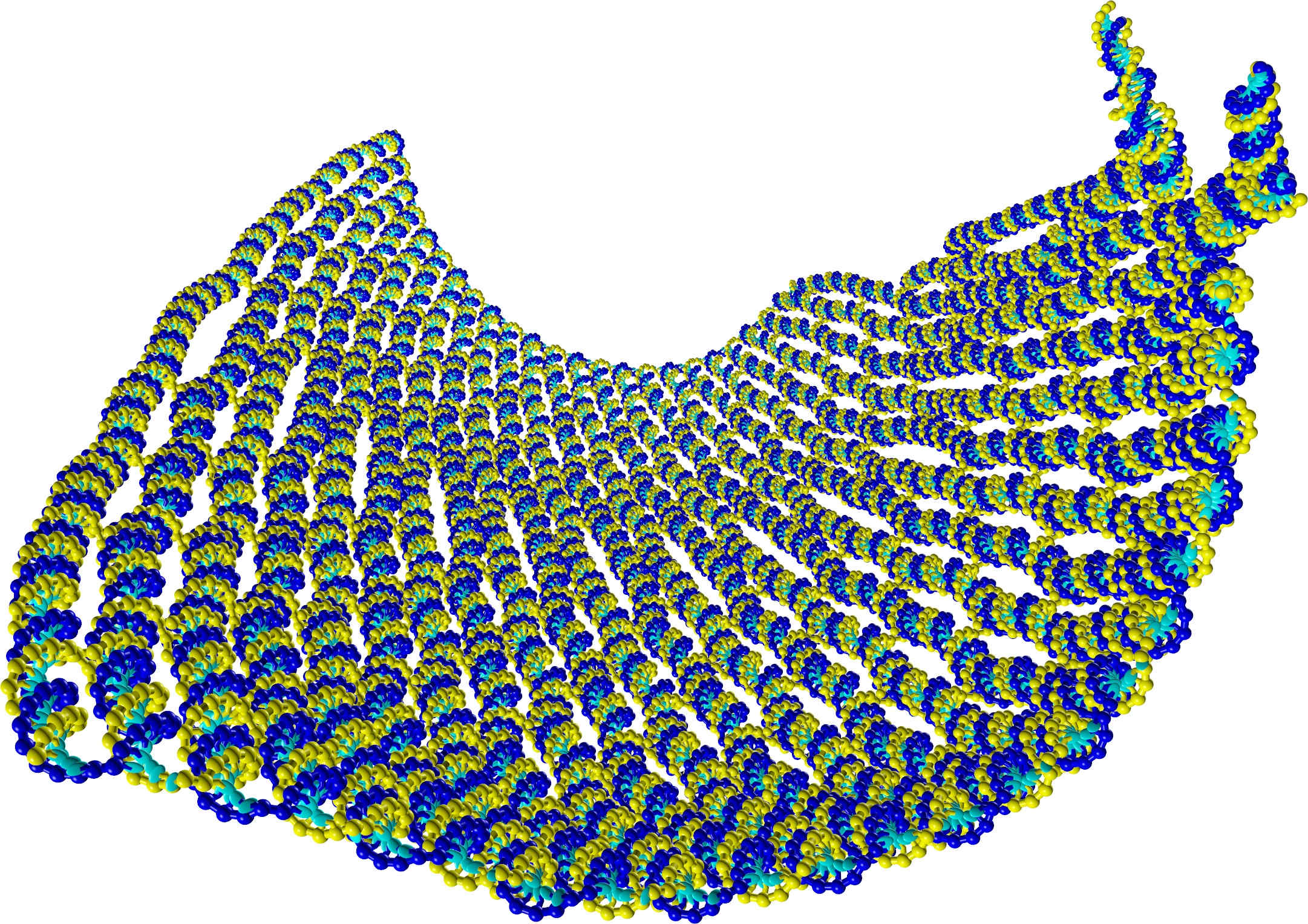
Abstract
We use the oxDNA coarse-grained model to provide a detailed characterization of the fundamental structural properties of DNA origamis, focussing on archetypal 2D and 3D origamis. The model reproduces well the characteristic pattern of helix bending in a 2D origami, showing that it stems from the intrinsic tendency of anti-parallel four-way junctions to splay apart, a tendency that is enhanced both by less screened electrostatic interactions and by increased thermal motion. We also compare to the structure of a 3D-origami whose structure has been determined by cryo-electron microscopy. The oxDNA average structure has a root-mean-square deviation from the experimental structure of 8.4 Angstrom which is of the order of the experimental resolution. These results illustrate that the oxDNA model is capable of providing detailed and accurate insights into the structure of DNA origamis, and has the potential to be used to routinely pre-screen putative origami designs.The full paper is available from Nucleic Acids Research and arXiv.org.