|
|
The role of loop stacking in the dynamics of DNA hairpin formation
Majid Mosayebi, Flavio Romano, Thomas E. Ouldridge, Ard A. Louis and Jonathan P.K. Doye
J. Phys. Chem. B 118, 14326-14335 (2014)
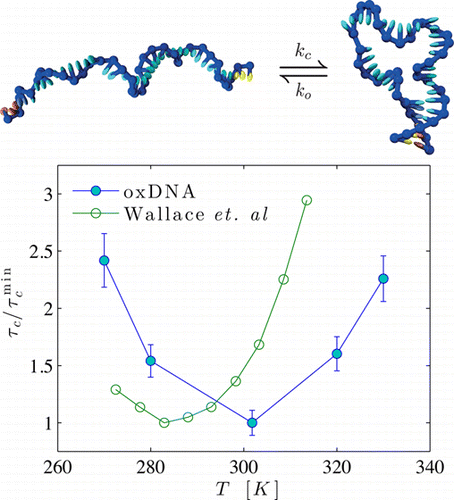
Abstract
We study the dynamics of DNA hairpin formation using oxDNA, a nucleotide-level coarse- grained model of DNA. In particular, we explore the effects of the loop stacking interactions and non-native base pairing on the hairpin closing times. We find a non-monotonic variation of the hairpin closing time with temperature, in agreement with the experimental work of Wallace et al. [Proc. Nat. Acad. Sci. USA 2001, 98 , 5584-5589]. The hairpin closing process involves the formation of an initial nucleus of one or two bonds between the stems followed by a rapid zippering of the stem. At high temperatures, typically above the hairpin melting temperature, an effective negative activation enthalpy is observed because the nucleus has a lower enthalpy than the open state. By contrast, at low temperatures, the activation enthalpy becomes positive mainly due to the increasing energetic cost of bending a loop that becomes increasingly highly stacked as the temperature decreases. We show that stacking must be very strong to induce this experimentally observed behavior, and that the existence of just a few weak stacking points along the loop can substantially suppress it. Non-native base pairs are observed to have only a small effect, slightly accelerating hairpin formation.The full paper is available from The Journal of Physical Chemistry B and arXiv.org.