|
|
DNA cruciform arms nucleate through a correlated but asynchronous cooperative mechanism
Christian Matek, Thomas E. Ouldridge, Adam Levy, Jonathan P.K. Doye and Ard A. Louis
J. Phys. Chem. B 116, 11616-11625 (2012)
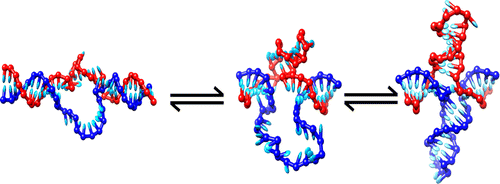
Abstract
Inverted repeat (IR) sequences in DNA can form non-canonical cruciform structures to relieve torsional stress. We use Monte Carlo simulations of a recently developed coarsegrained model of DNA to demonstrate that the nucleation of a cruciform can proceed through a cooperative mechanism. Firstly, a twist-induced denaturation bubble must diffuse so that its midpoint is near the centre of symmetry of the IR sequence. Secondly, bubble fluctuations must be large enough to allow one of the arms to form a small number of hairpin bonds. Once the first arm is partially formed, the second arm can rapidly grow to a similar size. Because bubbles can twist back on themselves, they need considerably fewer bases to resolve torsional stress than the final cruciform state does. The initially stabilised cruciform therefore continues to grow, which typically proceeds synchronously, reminiscent of the S-type mechanism of cruciform formation. By using umbrella sampling techniques we calculate, for different temperatures and superhelical densities, the free energy as a function of the number of bonds in each cruciform along the correlated but non-synchronous nucleation pathways we observed in direct simulationsThe full paper is available from The Journal of Physical Chemistry B and arXiv.org.