|
|
Computing the elastic mechanical properties of rod-like DNA nanostructures
Hemani Chhabra, Garima Mishra, Yijing Cao, Domen Prešern, Enrico Skoruppa, Maxime M.C. Tortora and Jonathan P.K. Doye
J. Chem. Theory Comput. 16, 7748–7763 (2020)
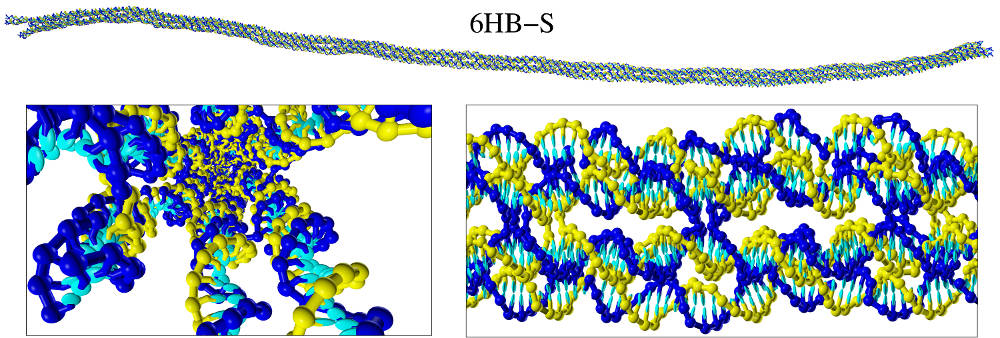
Abstract
To study the elastic properties of rod-like DNA nanostructures, we perform long simulations of these structure using the oxDNA coarse-grained model. By analysing the fluctuations in these trajectories we obtain estimates of the bend and twist persistence lengths, and the underlying bend and twist elastic moduli and couplings between them. Only on length scales beyond those associated with the spacings between the interhelix crossovers do the bending fluctuations behave like those of a worm-like chain. The obtained bending persistence lengths are much larger than that for double-stranded DNA and increase non-linearly with the number of helices, whereas the twist moduli increase approximately linearly. To within the numerical error in our data, the twist-bend coupling constants are of order zero. That the bending persistence lengths we obtain are generally somewhat higher than in experiment probably reflects both that the simulated origami have no assembly defects and that the oxDNA extensional modulus for double-stranded DNA is too large.The full paper is available from J. Chem. Theory Comput. and arXiv.org.