|
|
Identifying physical causes of apparent enhanced cyclization of short DNA molecules with a coarse-grained model
Ryan M. Harrison, Flavio Romano, Thomas E. Ouldridge, Ard A. Louis and Jonathan P.K. Doye
J. Chem. Theory Comput. 15, 4660-4672 (2019)
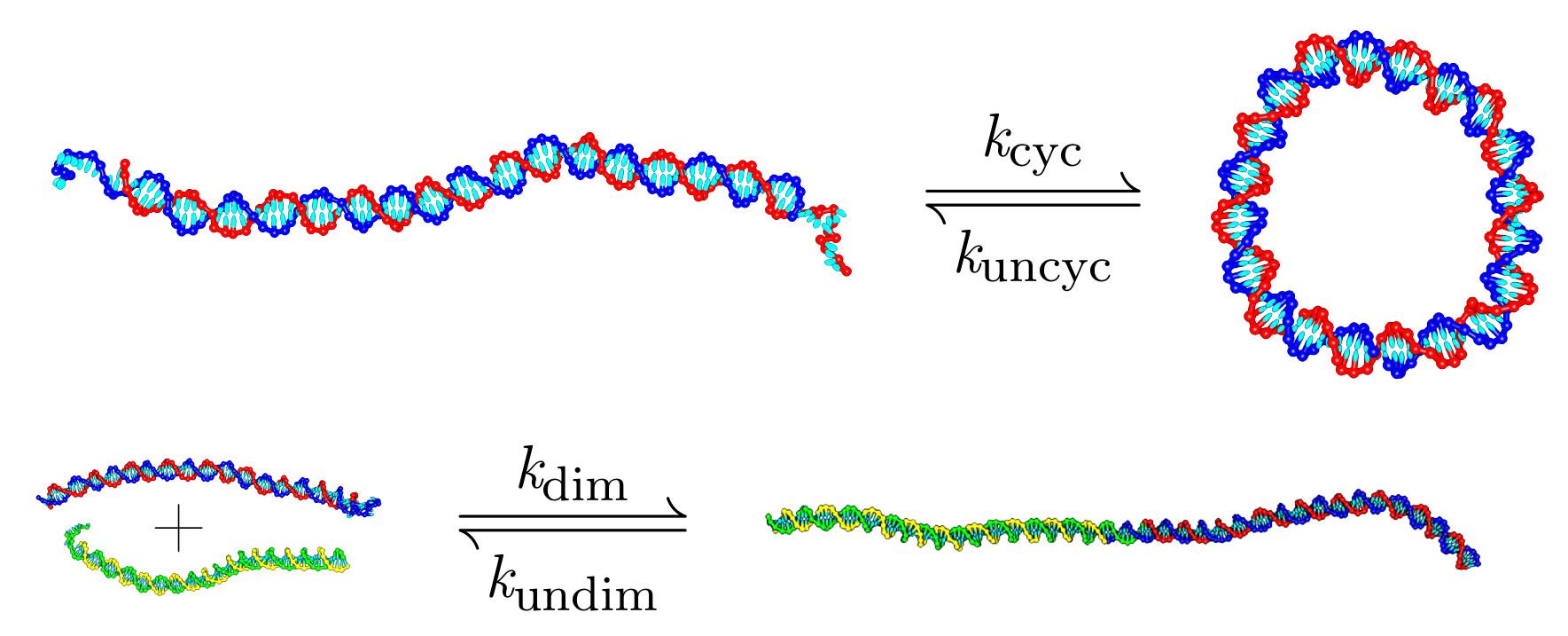
Abstract
DNA cyclization is a powerful technique to gain insight into the nature of DNA bending. The worm-like chain model provides a good description of small to moderate bending fluctuations, but some experiments on strongly-bent shorter molecules suggest enhanced flexibility over and above that expected from the worm-like chain. Here, we use a coarse-grained model of DNA to investigate the thermodynamics of DNA cyclization for molecules with less than 210 base pairs. As the molecules get shorter we find increasing deviations between our computed equilibrium j-factor and the worm-like chain predictions of Shimada and Yamakawa. These deviations are due to sharp kinking, first at nicks, and only subsequently in the body of the duplex. At the shortest lengths, substantial fraying at the ends of duplex domains is the dominant method of relaxation. We also estimate the dynamic j-factor measured in recent FRET experiments. We find that the dynamic j-factor is systematically larger than its equilibrium counterpart, with the deviation larger for shorter molecules, because not all the stress present in the fully cyclized state is present in the transition state. These observations are important for the interpretation of recent experiments, as only kinking within the body of the duplex is genuinely indicative of non-worm-like chain behaviour.The full paper is available from the Journal of Chemical Theory and Computation and arXiv.org.