|
|
Force-induced unravelling of DNA Origami
Megan C. Engel, David M. Smith, Markus A. Jobst, Martin Sajfutdinow, Tim Liedl, Flavio Romano, Lorenzo Rovigatti, Ard A. Louis and Jonathan P.K. Doye
ACS Nano, 12, 6734-6747 (2018)
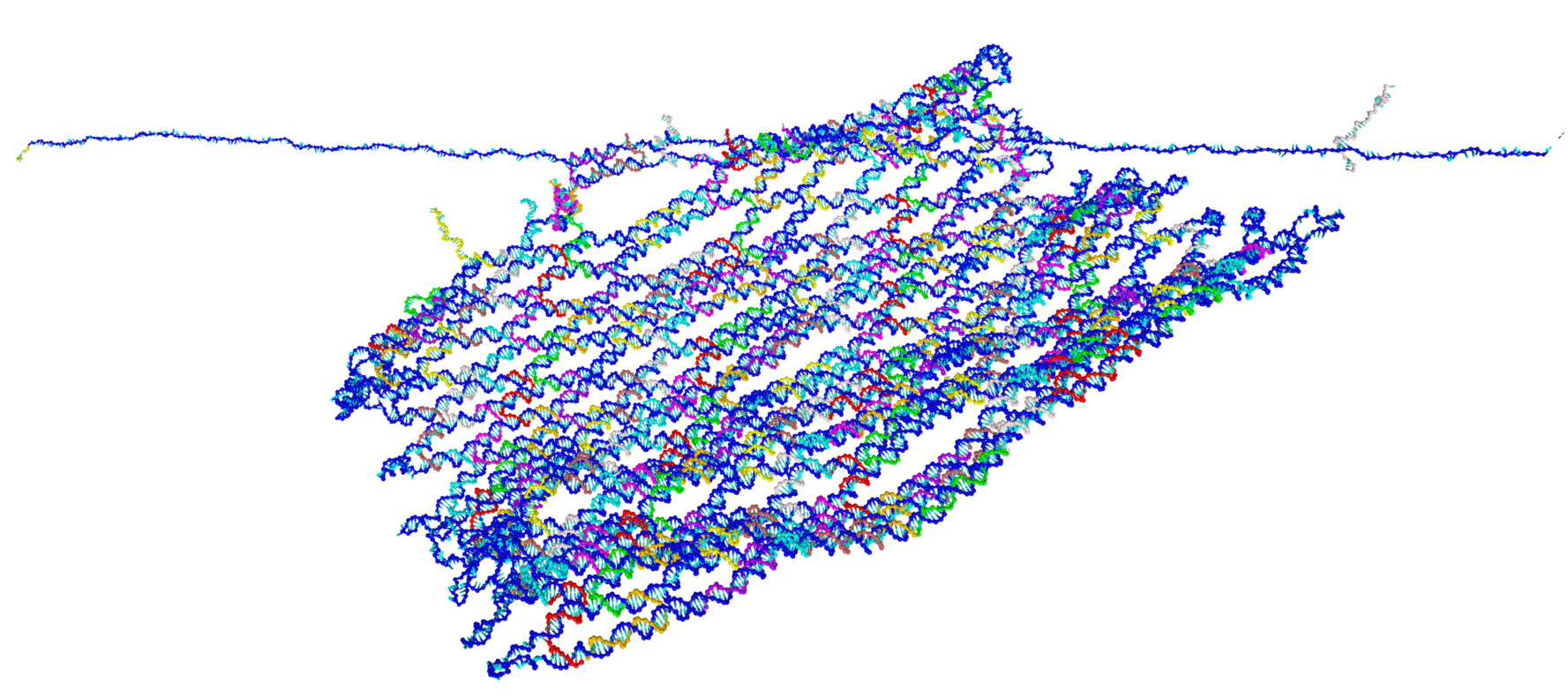
Abstract
The mechanical properties of DNA nanostructures are of widespread interest as applications that exploit their stability under constant or intermittent external forces become increasingly common. We explore the force response of DNA origami in unprecedented detail by combining AFM single molecule force spectroscopy experiments with simulations using oxDNA, a coarsegrained model of DNA at the nucleotide level, to study the unravelling of an iconic origami system: the Rothemund tile. We contrast the force-induced melting of the tile with simulations of an origami 10-helix bundle. Finally, we simulate a recently-proposed origami biosensor, whose function takes advantage of origami behaviour under tension. We observe characteristic stick-slip unfolding dynamics in our forceextension curves for both the Rothemund tile and the helix bundle and reasonable agreement with experimentally observed rupture forces for these systems. Our results highlight the ect of design on force response: we observe regular, modular unfolding for the Rothemund tile that contrasts with strain-softening of the 10- helix bundle which leads to catastropic failure under monotonically increasing force. Further, unravelling occurs straightforwardly from the scald ends inwards for the Rothemund tile, while the helix bundle unfolds more nonlinearly. The detailed visualization of the yielding events provided by simulation allows preferred pathways through the complex unfolding free-energy landscape to be mapped, as a key factor in determining relative barrier heights is the extensional release per base pair broken. We shed light on two important questions: how stable DNA nanostructures are under external forces; and what design principles can be applied to enhance stability.The full paper is available from ACS Nano