|
|
Direct simulation of the self-assembly of a small DNA origami
Benedict E. K. Snodin, Flavio Romano, L. Rovigatti, Thomas E. Ouldridge, Ard A. Louis and Jonathan P.K. Doye
ACS Nano, 10, 1724-1737 (2016)
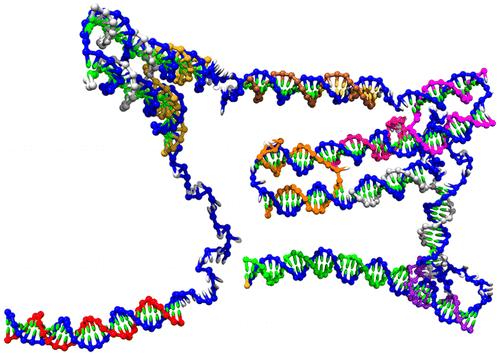
Abstract
By using oxDNA, a coarse-grained nucleotide-level model of DNA, we are able to directly simulate the self-assembly of a small 384-base-pair origami from single-stranded scald and staple strands in solution. In general, we see attachment of new staple strands occurring in parallel, but with cooperativity evident for the binding of the second domain of a staple if the adjacent junction is already partially formed. For a system with exactly one copy of each staple strand, we observe a complete assembly pathway in an intermediate temperature window; at low temperatures successful assembly is prevented by misbonding while at higher temperature the free-energy barriers to assembly become too large for assembly on our simulation time scales. For high-concentration systems involving a large staple strand excess, we never see complete assembly because there are invariably instances where copies of the same staple both bind to the scald, creating a kinetic trap that prevents the complete binding of either staple. This mutual staple blocking could also lead to aggregates of partially formed origamis in real systems, and helps to rationalize certain successful origami design strategies.The full paper is available from ACS Nano and for non-subscribers from ACS articles on request.